IPT Transition at crossover¶
Study the end of the crossover region
from __future__ import division, absolute_import, print_function
from dmft.ipt_imag import dmft_loop
from dmft.common import greenF, tau_wn_setup, fit_gf
from dmft.twosite import matsubara_Z
import numpy as np
import matplotlib.pylab as plt
def hysteresis(beta, u_range):
log_g_sig = []
tau, w_n = tau_wn_setup(dict(BETA=beta, N_MATSUBARA=beta))
g_iwn = greenF(w_n)
for u_int in u_range:
g_iwn, sigma = dmft_loop(u_int, 0.5, g_iwn, w_n, tau)
log_g_sig.append((g_iwn, sigma))
return log_g_sig
U = np.linspace(2.2, 2.5, 100)
results = []
betarange = [16, 17.85, 19.2, 20., 21.3]
for beta in betarange:
results.append(hysteresis(beta, U))
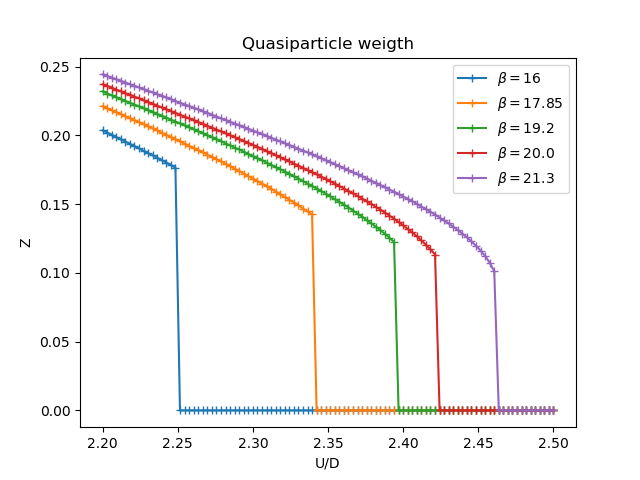
Quasiparticle weight¶
figz, axz = plt.subplots()
for beta, result in zip(betarange, results):
u_zet = [matsubara_Z(sigma.imag, beta) for _, sigma in result]
axz.plot(U, u_zet, '+-', label='$\\beta={}$'.format(beta))
axz.set_title('Quasiparticle weigth')
axz.legend(loc=0)
axz.set_ylabel('Z')
axz.set_xlabel('U/D')
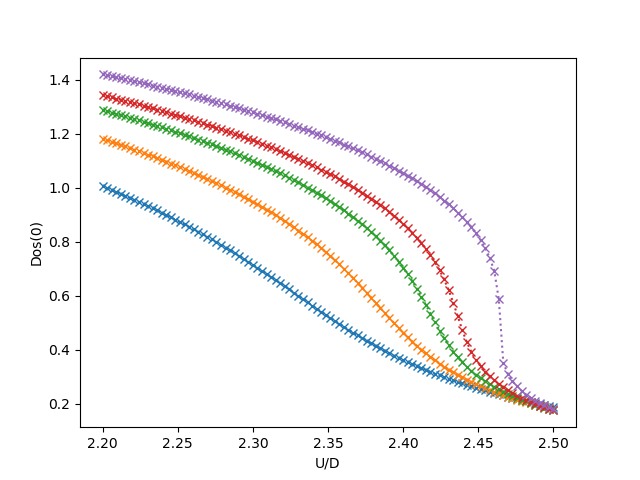
Spectral density at Fermi level¶
figf, axf = plt.subplots()
for beta, result in zip(betarange, results):
tau, w_n = tau_wn_setup(dict(BETA=beta, N_MATSUBARA=3))
u_fl = [-fit_gf(w_n, g_iwn.imag)(0.)for g_iwn, _ in result]
axf.plot(U, u_fl, 'x:', label='$\\beta={}$'.format(beta))
axf.set_ylabel('Dos(0)')
axz.set_title('Density of states at Fermi level')
axf.set_xlabel('U/D')
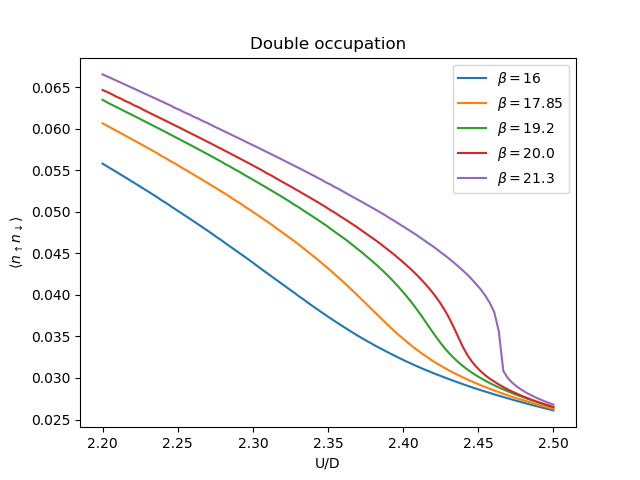
Double occupation¶
Is proportional to the Potential energy which is defined in Potential energy the tail of this function behaves as
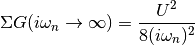
Then to find out the double occupation one uses the relation
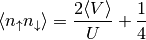
figd, axd = plt.subplots()
for beta, result in zip(betarange, results):
tau, w_n = tau_wn_setup(dict(BETA=beta, N_MATSUBARA=beta))
V = np.asarray([2 * (0.5 * s * g + u**2 / 8. / w_n**2).real.sum() / beta
for (g, s), u in zip(result, U)]) - 0.25 * beta * U**2 / 8.
D = 2. / U * V + 0.25
axd.plot(U, D, '-', label='$\\beta={}$'.format(beta))
axd.set_title('Double occupation')
axd.legend(loc=0)
axd.set_ylabel(r'$\langle n_\uparrow n_\downarrow \rangle$')
axd.set_xlabel('U/D')
Total running time of the script: ( 0 minutes 1.452 seconds)