Landau Theory of the Mott transition¶
Perform a fit of the order parameter, linked to double occupation to match a Landau theory formulation in correspondence to Kotliar, G., Lange, E., & Rozenberg, M. J. (2000). Landau Theory of the Finite Temperature Mott Transition. Phys. Rev. Lett., 84(22), 5180–5183. http://dx.doi.org/10.1103/PhysRevLett.84.5180
Study above the critical point
Out:
[67 61 62 61 59 57 55 53 51 49 47 45 44 42 41 39 38 37 37 37 38 38 38 39 39
39 39 39 39 40 40 40 40 40 40 40 40 41 41 41 41 41 41 41 41 41 41 42 42 42
42 42 42 42 42 42 42 42 43 43 43 43 43 43 43 43 43 43 44 44 44 44 44 44 44
44 44 44 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 45 44
44 44 43 43 42 42 41 40 39 38 37 36 35 33 32 30 28 27 25 23 21]
[68 64 66 67 66 65 64 62 60 58 56 54 52 50 49 47 46 44 43 42 42 43 43 44 44
45 45 46 46 47 47 48 48 49 49 49 50 50 51 51 51 52 52 53 53 53 54 54 54 55
55 55 56 56 56 57 57 57 58 58 58 59 59 59 59 60 60 60 60 61 61 61 61 61 62
62 62 62 62 62 62 62 62 62 61 61 61 61 60 60 59 59 58 57 57 56 55 54 53 52
51 49 48 47 45 44 42 41 39 38 36 34 33 31 29 28 26 24 22 21 19]
[67 64 68 70 71 71 71 70 68 67 65 63 61 59 58 56 54 53 51 50 48 47 47 49 49
50 51 52 53 54 55 56 57 58 60 61 62 63 64 65 66 67 68 70 71 72 73 75 76 77
78 80 81 82 83 85 86 87 88 89 90 91 92 93 94 95 95 95 96 96 95 95 94 94 93
92 91 89 88 86 84 83 81 79 77 75 73 71 69 67 65 63 61 59 57 55 53 51 50 48
46 45 43 41 40 38 37 35 34 32 31 30 28 27 25 24 23 21 20 18 17]
[ 67 64 68 71 72 73 73 73 72 71 69 68 66 64 63 61 59 58
56 54 53 51 50 49 51 52 53 54 56 57 58 60 61 62 64 66
67 69 70 72 74 76 78 80 82 84 86 89 91 94 97 99 102 105
108 111 113 116 119 122 124 126 127 128 129 129 129 127 126 124 121 118
115 111 108 104 100 97 93 89 86 83 80 77 74 71 69 66 64 61
59 57 55 53 51 50 48 46 45 43 42 40 39 37 36 35 33 32
31 30 28 27 26 25 24 22 21 20 19 17 16]
[ 65 63 67 70 73 74 75 76 75 75 74 72 71 70 68 67 65 63
62 60 59 57 56 54 53 52 53 54 55 57 58 60 61 63 65 67
69 71 73 75 78 80 83 86 89 92 96 100 104 109 114 120 126 132
139 147 155 164 172 180 187 193 195 195 192 185 177 168 158 148 139 130
122 114 108 102 96 91 87 83 79 75 72 69 66 63 61 59 57 55
53 51 49 47 46 44 43 41 40 39 37 36 35 34 33 32 31 29
28 27 26 25 24 23 22 21 20 19 18 17 15]
[ 63 61 65 69 72 74 76 77 77 77 77 76 75 74 73 72 71 69
68 66 65 64 62 61 60 58 57 56 55 55 56 57 59 61 63 65
66 69 71 73 76 79 82 85 88 92 97 102 107 113 120 129 139 150
164 182 204 233 270 316 364 387 363 313 262 222 191 168 149 135 123 113
105 98 92 86 81 77 73 70 67 64 62 59 57 55 53 51 49 48
46 45 43 42 41 39 38 37 36 35 34 33 32 31 30 29 28 27
26 25 24 24 23 22 21 20 19 18 17 16 15]
[ 63 60 65 69 72 74 76 77 78 78 78 78 77 76 75 74 73 71
70 69 67 66 65 63 62 61 60 58 57 56 55 57 58 60 61 63
65 67 69 72 74 77 80 83 87 91 95 100 106 112 120 129 140 153
170 191 222 266 336 454 604 554 393 290 229 191 164 144 129 118 108 100
93 87 82 78 74 70 67 64 62 59 57 55 53 51 49 48 46 45
44 42 41 40 39 38 36 35 34 33 33 32 31 30 29 28 27 26
25 25 24 23 22 21 20 19 19 18 17 16 15]
[ -3.86528847e+04 -2.47024433e+01 1.76230198e+01 -6.76142889e-04]
[ -5.66140107e+04 -1.90645920e+01 6.03415912e+00 -1.03353073e-04]
[ -9.29427951e+04 -1.32890339e+01 1.65847125e+00 -2.43110956e-04]
[ -1.25002557e+05 -1.01922703e+01 -3.05401520e-01 -1.92401678e-04]
[ -1.58468430e+05 -7.01333087e+00 1.42348154e+00 -3.21099956e-04]
[ -1.93447631e+05 -3.76601834e+00 9.68173850e+00 -5.89656130e-04]
[ -2.14218382e+05 -2.43545432e+00 4.22759934e+01 -4.49653695e-04]
from scipy.optimize import curve_fit
import numpy as np
import matplotlib.pyplot as plt
import dmft.dimer as dimer
import dmft.common as gf
import dmft.ipt_imag as ipt
def loop_u_tp(u_range, tprange, beta, seed='mott gap'):
tau, w_n = gf.tau_wn_setup(dict(BETA=beta, N_MATSUBARA=256))
giw_d, giw_o = dimer.gf_met(w_n, 0., 0., 0.5, 0.)
if seed == 'ins':
giw_d, giw_o = 1 / (1j * w_n + 4j / w_n), np.zeros_like(w_n) + 0j
giw_s = []
sigma_iw = []
ekin, epot = [], []
iterations = []
for u_int, tp in zip(u_range, tprange):
giw_d, giw_o, loops = dimer.ipt_dmft_loop(
beta, u_int, tp, giw_d, giw_o, tau, w_n)
giw_s.append((giw_d, giw_o))
iterations.append(loops)
g0iw_d, g0iw_o = dimer.self_consistency(
1j * w_n, 1j * giw_d.imag, giw_o.real, 0., tp, 0.25)
siw_d, siw_o = ipt.dimer_sigma(u_int, tp, g0iw_d, g0iw_o, tau, w_n)
sigma_iw.append((siw_d.copy(), siw_o.copy()))
ekin.append(dimer.ekin(giw_d, giw_o, w_n, tp, beta))
epot.append(dimer.epot(giw_d, w_n, beta, u_int **
2 / 4 + tp**2, ekin[-1], u_int))
print(np.array(iterations))
# last division in energies because I want per spin epot
return np.array(giw_s), np.array(sigma_iw), np.array(ekin) / 4, np.array(epot) / 4, w_n
# calculating multiple regions
fac = np.arctan(.55 * np.sqrt(3) / .15)
udelta = np.tan(np.linspace(-fac, fac, 121)) * .15 / np.sqrt(3)
dudelta = np.diff(udelta)
bet_uc = [(18, 3.312),
(19, 3.258),
(20, 3.214),
(20.5, 3.193),
(21, 3.17),
(21.5, 3.1467),
(21.7, 3.138)]
data = []
for beta, uc in bet_uc:
urange = udelta + uc + .07
giw_s, sigma_iw, ekin, epot, w_n = loop_u_tp(
urange, .3 * np.ones_like(urange), beta, 'met')
data.append(2 * epot / urange - 0.003)
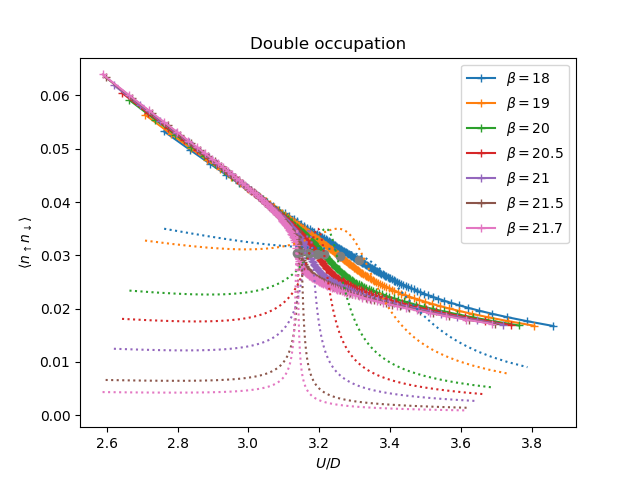
plt.figure()
bc = [b for b, _ in bet_uc]
d_c = [dc[int(len(udelta) / 2)] for dc in data]
for dd, dc, (beta, uc) in zip(data, d_c, bet_uc):
plt.plot(uc + udelta, dd, '+-', label=r'$\beta={}$'.format(beta))
plt.plot([uc for _, uc in bet_uc], d_c, 'o')
plt.gca().set_color_cycle(None)
for dd, (beta, uc) in zip(data, bet_uc):
chi = np.diff(dd) / dudelta
plt.plot(uc + udelta[:-1], chi / np.min(chi) * .035, ':')
plt.title(r'Double occupation')
plt.ylabel(r'$\langle n_\uparrow n_\downarrow \rangle$')
plt.xlabel(r'$U/D$')
plt.legend()
plt.savefig("dimer_tp0.3_docc.pdf",
transparent=False, bbox_inches='tight', pad_inches=0.05)
# effective scaling
# cubic + linear over constant + linear
def fit_cube_lin(eta, c, p, q, s):
return (c * eta**3 + p * eta + s) / (1 + q * eta)
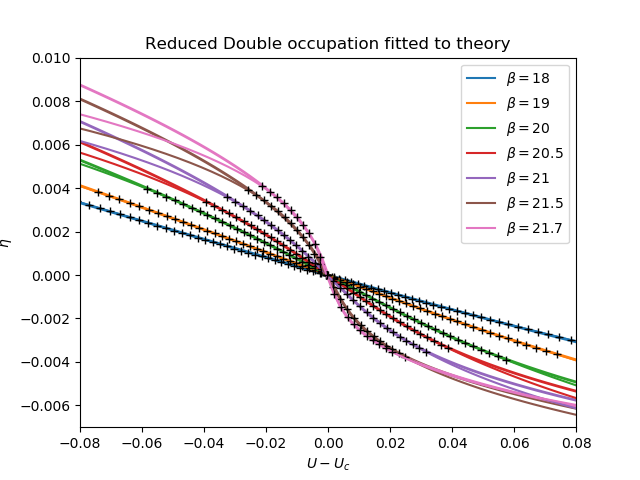
plt.figure()
for dd, dc, (beta, uc) in zip(data, d_c, bet_uc):
plt.plot(udelta, dd - dc, lw=2)
plt.gca().set_color_cycle(None)
bb = [10, 30, 35, 42, 45, 48, 50]
fits = []
for dd, dc, bound, (beta, uc) in zip(data, d_c, bb, bet_uc):
rd = dd - dc
popt, pcov = curve_fit(
fit_cube_lin, rd[bound:-bound], udelta[bound:-bound], p0=[-4e4, -3, 3, 1])
fits.append((popt, pcov))
ft = fit_cube_lin(rd, *popt)
plt.plot(ft, rd, label=r'$\beta={}$'.format(beta))
plt.plot(ft[bound:-bound], rd[bound:-bound], "k+")
print(popt)
plt.xlim([-.08, .08])
plt.ylim([-.007, .01])
plt.title(r'Reduced Double occupation fitted to theory')
plt.ylabel(r'$\eta$')
plt.xlabel(r'$U-U_c$')
plt.legend()
plt.savefig("dimer_tp0.3_eta_landau.pdf",
transparent=False, bbox_inches='tight', pad_inches=0.05)
Total running time of the script: ( 1 minutes 17.737 seconds)